import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import statsmodels.formula.api as smf1 Régression simple
La concentration en ozone
ozone = pd.read_csv("../donnees/ozone_simple.txt", header=0, sep=";")
plt.plot(ozone.T12, ozone.O3, '.k')
plt.ylabel('O3')
plt.xlabel('T12')
plt.show()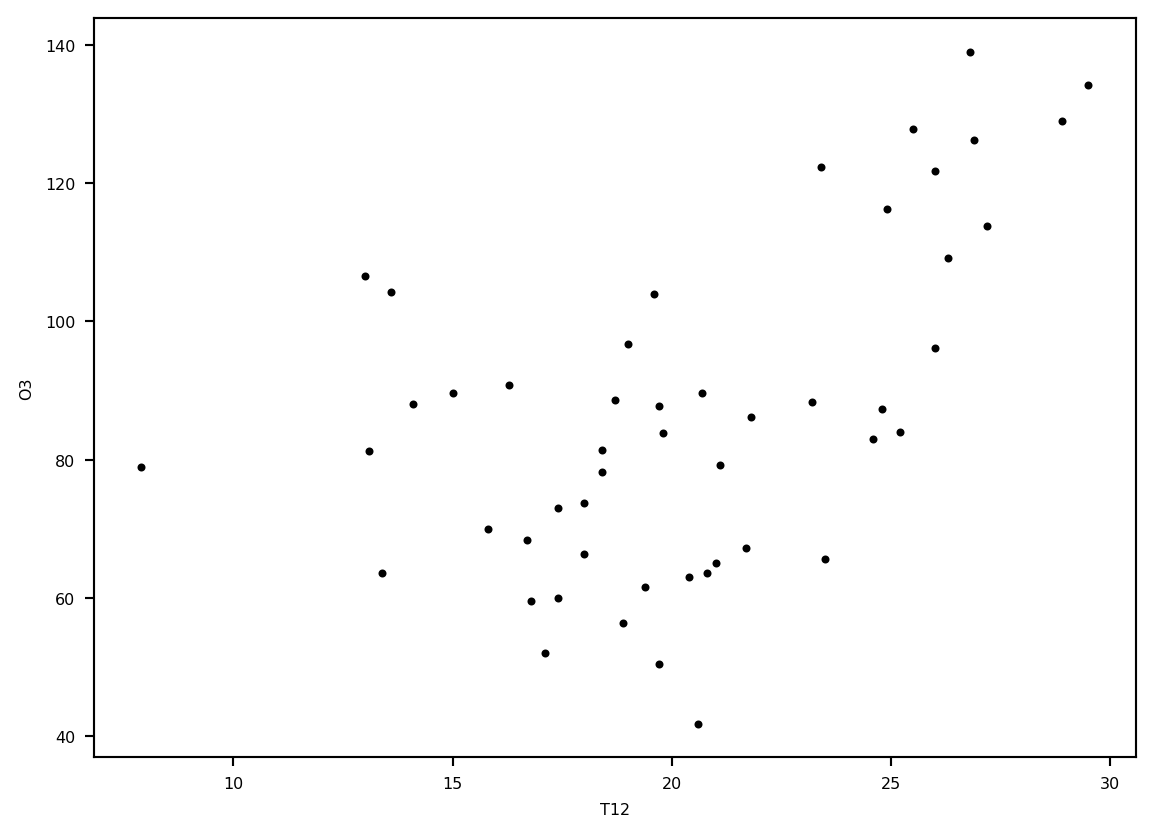
reg = smf.ols('O3 ~ T12', data=ozone).fit()
reg.summary()| Dep. Variable: | O3 | R-squared: | 0.279 |
| Model: | OLS | Adj. R-squared: | 0.264 |
| Method: | Least Squares | F-statistic: | 18.58 |
| Date: | Fri, 31 Jan 2025 | Prob (F-statistic): | 8.04e-05 |
| Time: | 15:41:21 | Log-Likelihood: | -220.96 |
| No. Observations: | 50 | AIC: | 445.9 |
| Df Residuals: | 48 | BIC: | 449.7 |
| Df Model: | 1 | ||
| Covariance Type: | nonrobust |
| coef | std err | t | P>|t| | [0.025 | 0.975] | |
| Intercept | 31.4150 | 13.058 | 2.406 | 0.020 | 5.159 | 57.671 |
| T12 | 2.7010 | 0.627 | 4.311 | 0.000 | 1.441 | 3.961 |
| Omnibus: | 2.252 | Durbin-Watson: | 1.461 |
| Prob(Omnibus): | 0.324 | Jarque-Bera (JB): | 1.348 |
| Skew: | 0.026 | Prob(JB): | 0.510 |
| Kurtosis: | 2.197 | Cond. No. | 94.1 |
Notes:
[1] Standard Errors assume that the covariance matrix of the errors is correctly specified.
grille = pd.DataFrame({'T12': np.linspace(ozone.T12.min(), ozone.T12.max(), 100)})
calculprev = reg.get_prediction(grille)
ICdte = calculprev.conf_int(obs=False, alpha=0.05)
prev = calculprev.predicted_mean
ICprev = calculprev.conf_int(obs=True, alpha=0.05)plt.plot(ozone.T12, ozone.O3, '.k')
plt.ylabel('O3')
plt.xlabel('T12')
plt.plot(grille.T12, prev, '-', lw=1, label="E(Y)")
lesic, = plt.plot(grille.T12, ICdte[:, 0], 'r--', label=r"IC $\mathbb{E}(Y)$", lw=1)
plt.plot(grille.T12, ICdte[:, 1], 'r--', lw=1)
plt.legend(handles=[lesic,], loc='lower right')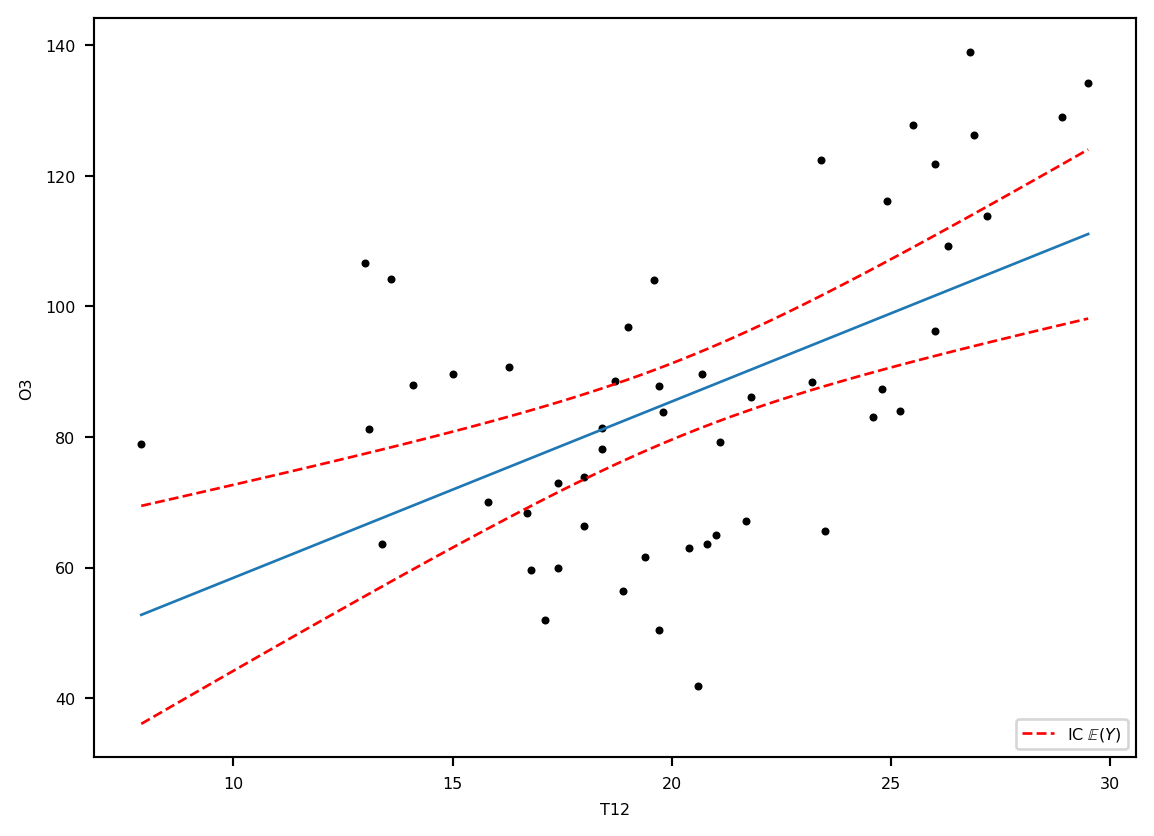
plt.plot(ozone.T12, ozone.O3, '.k')
plt.ylabel('O3') ; plt.xlabel('T12')
plt.plot(grille.T12, prev, '-', lw=1, label="E(Y)")
lesic, = plt.plot(grille.T12, ICdte[:, 0], 'r--', label=r"IC $\mathbb{E}(Y)$", lw=1)
plt.plot(grille.T12, ICdte[:, 1],'r--', lw=1)
lesic2, = plt.plot(grille.T12, ICprev[:, 0], 'g:', label=r"IC $Y$", lw=1)
plt.plot(grille.T12, ICprev[:, 1],'g:', lw=1)
plt.legend(handles=[lesic, lesic2], loc='lower right')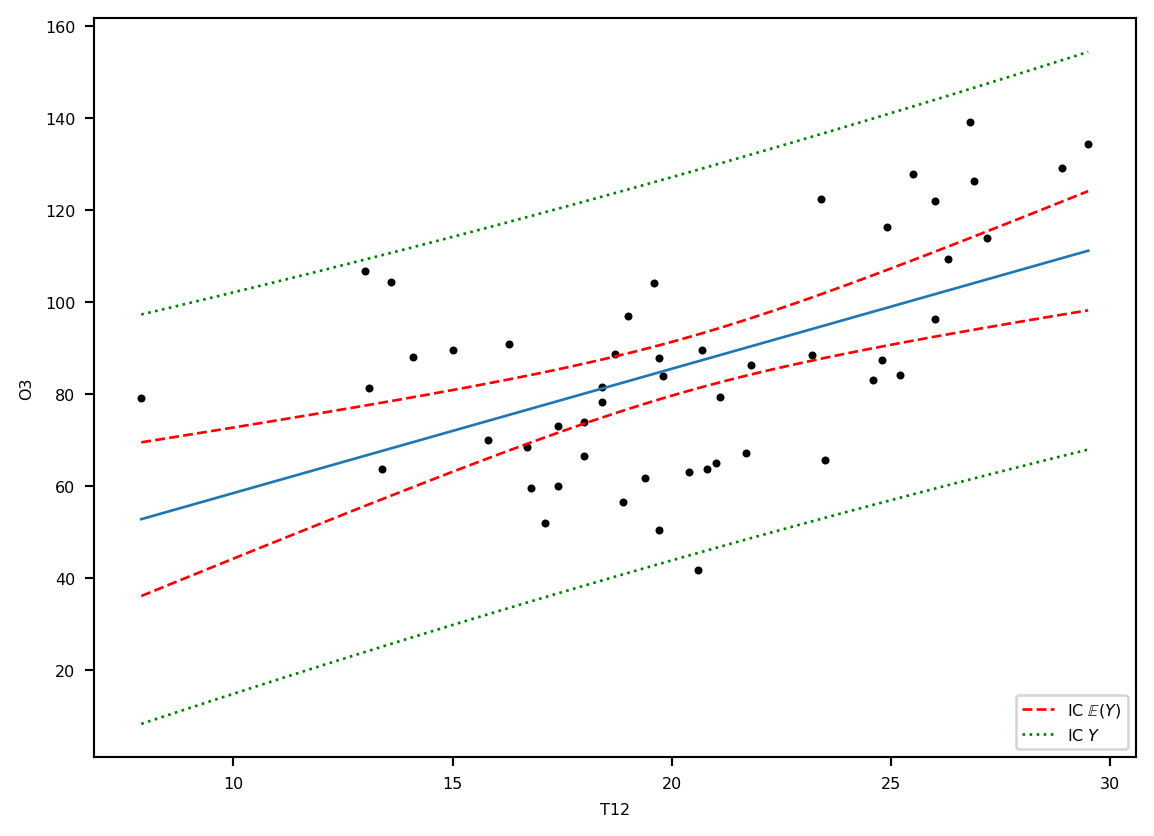
ICparams = reg.conf_int(alpha=0.05)
round(ICparams, 3)| 0 | 1 | |
|---|---|---|
| Intercept | 5.159 | 57.671 |
| T12 | 1.441 | 3.961 |
La hauteur des eucalyptus
eucalyptus = pd.read_csv("../donnees/eucalyptus.txt", header=0, sep=";")
plt.plot(eucalyptus.circ, eucalyptus.ht, '+k')
plt.ylabel('ht')
plt.xlabel('circ')
plt.show()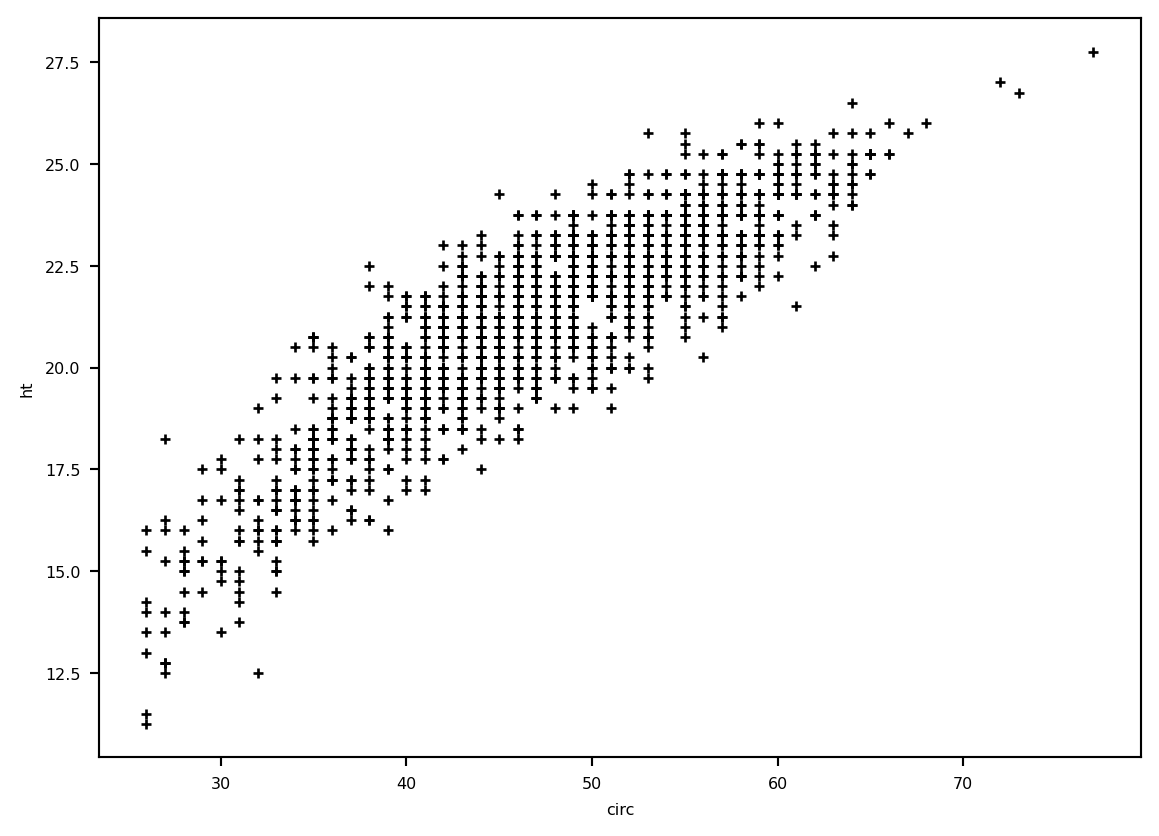
reg = smf.ols('ht ~ circ', data=eucalyptus).fit()
reg.summary()| Dep. Variable: | ht | R-squared: | 0.768 |
| Model: | OLS | Adj. R-squared: | 0.768 |
| Method: | Least Squares | F-statistic: | 4732. |
| Date: | Fri, 31 Jan 2025 | Prob (F-statistic): | 0.00 |
| Time: | 15:41:22 | Log-Likelihood: | -2286.2 |
| No. Observations: | 1429 | AIC: | 4576. |
| Df Residuals: | 1427 | BIC: | 4587. |
| Df Model: | 1 | ||
| Covariance Type: | nonrobust |
| coef | std err | t | P>|t| | [0.025 | 0.975] | |
| Intercept | 9.0375 | 0.180 | 50.264 | 0.000 | 8.685 | 9.390 |
| circ | 0.2571 | 0.004 | 68.792 | 0.000 | 0.250 | 0.264 |
| Omnibus: | 7.943 | Durbin-Watson: | 1.067 |
| Prob(Omnibus): | 0.019 | Jarque-Bera (JB): | 8.015 |
| Skew: | -0.156 | Prob(JB): | 0.0182 |
| Kurtosis: | 3.193 | Cond. No. | 273. |
Notes:
[1] Standard Errors assume that the covariance matrix of the errors is correctly specified.
grille = pd.DataFrame({'circ' : np.linspace(eucalyptus.circ.min(), eucalyptus.circ.max(), 100)})
calculprev = reg.get_prediction(grille)
ICdte = calculprev.conf_int(obs=False, alpha=0.05)
ICprev = calculprev.conf_int(obs=True, alpha=0.05)
prev = calculprev.predicted_meanplt.plot(eucalyptus.circ, eucalyptus.ht, '+k')
plt.ylabel('ht') ; plt.xlabel('circ')
plt.plot(grille.circ, prev, '-', label="E(Y)", lw=1)
lesic, = plt.plot(grille.circ, ICdte[:, 0], 'r--', label=r"IC $\mathbb{E}(Y)$", lw=1)
plt.plot(grille.circ, ICdte[:, 1], 'r--', lw=1)
plt.legend(handles=[lesic], loc='lower right')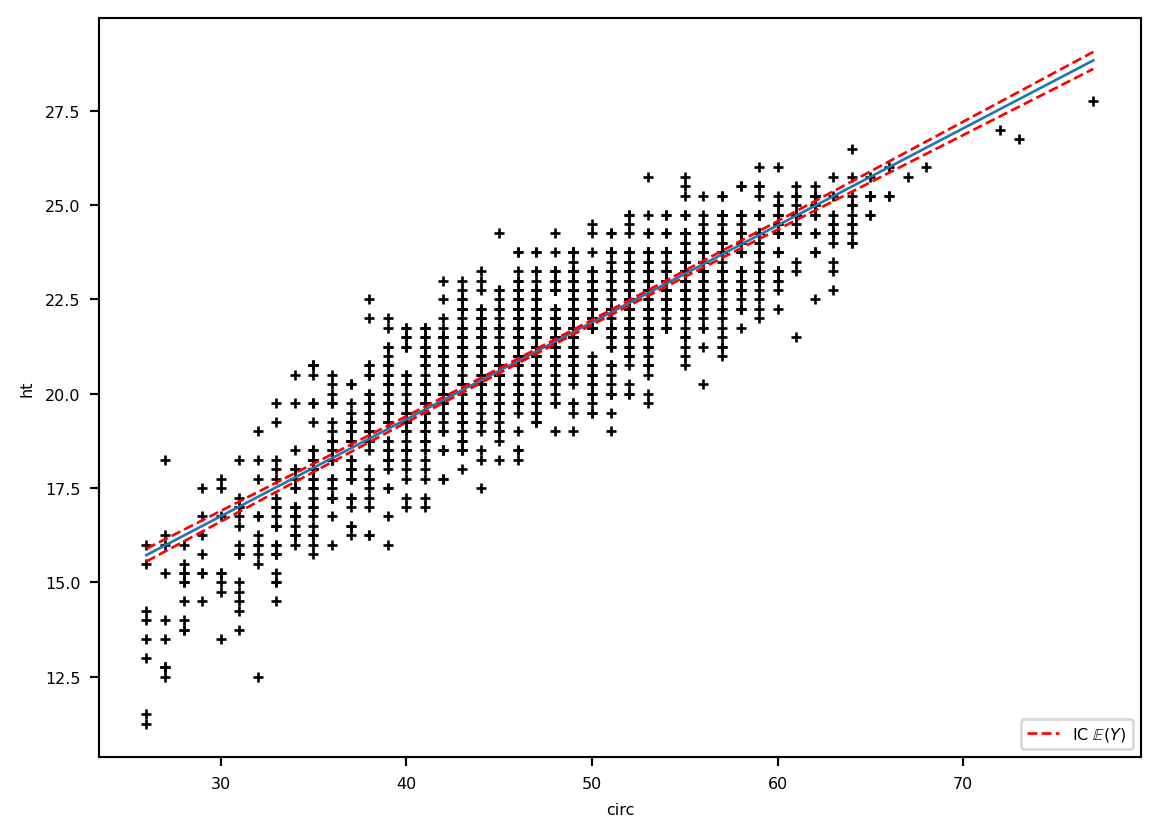
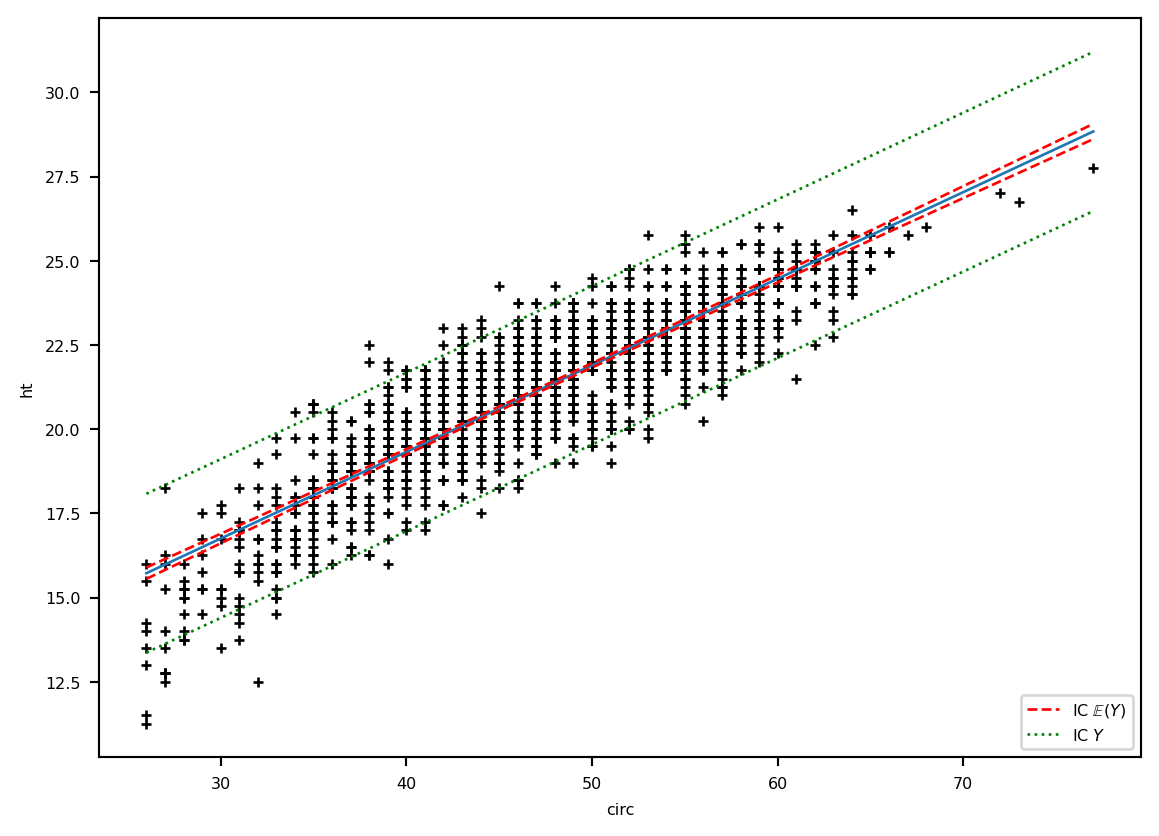
plt.plot(eucalyptus.circ, eucalyptus.ht, '+k')
plt.ylabel('ht') ; plt.xlabel('circ')
plt.plot(grille.circ, prev, '-', label="E(Y)", lw=1)
lesic, = plt.plot(grille.circ, ICdte[:, 0], 'r--', label=r"IC $\mathbb{E}(Y)$", lw=1)
plt.plot(grille.circ, ICdte[:, 1], 'r--', lw=1)
lesic2, = plt.plot(grille.circ, ICprev[:, 0], 'g:', label=r"IC $Y$", lw=1)
plt.plot(grille.circ, ICprev[:, 1],'g:', lw=1)
plt.legend(handles=[lesic, lesic2], loc='lower right')